function() {#:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# Intro to the Tidyverse by Colleen O'Briant
# Koan #15: map()
#:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# In order to progress:
# 1. Read all instructions carefully.
# 2. When you come to an exercise, fill in the blank, un-comment the line
# (Ctrl/Cmd Shift C), and execute the code in the console (Ctrl/Cmd Return).
# If the piece of code spans multiple lines, highlight the whole chunk or
# simply put your cursor at the end of the last line.
# 3. Save (Ctrl/Cmd S).
# 4. Test that your answers are correct (Ctrl/Cmd Shift T).
#:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# In this class, we've learned a lot about two very important tidyverse
# packages: 'dplyr' and 'ggplot2'. We'll spend the rest of the course
# learning a third: 'purrr'. We'll only learn 3 functions from that
# package, but they are deep concepts and really powerful tools. The
# three functions are map(), reduce(), and accumulate().
# This koan introduces map(), the next one gives you more practice using
# it, and reduce() and accumulate() will come a little later in koans
# 18-20.
# Run this code to get started:
library(tidyverse)
library(gapminder)
#:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# ----- Vectorized Functions -----
# Last week we learned we could define our own custom functions. Here I
# define a function called 'pct_change' that takes two arguments: an "old"
# value and a "new" value, and it returns the percentage change between
# them.
pct_change <- function(old, new) {
(new - old) / old
}
# 1. Does 'pct_change' work on values? ---------------------------------------
#1@
# pct_change(__, __) == 1
#@1
# 2. Does 'pct_change' work on vectors? --------------------------------------
#2@
# pct_change(__, __) == c(0, 1, 2)
#@2
# How is it that 'pct_change' works on vectors? Well, 'pct_change' is defined
# only using subtraction and division, which both work element-wise on
# vectors. You can plug vectors directly into the computation that's done in
# the function body:
(c(4, 5, 6) - c(1, 2, 3)) / c(1, 2, 3)
# ----- Example 1: rnorm -----
# Most of the functions we've been using are "vectorized" (they work with
# vectors of any length). But some functions may not work on vectors so
# smoothly. And when you define your own custom functions that are more
# complicated than `pct_change`, they oftentimes won’t work on vectors.
# For example, take 'rnorm()'.
# 3. Use 'rnorm()' to generate 10 random numbers from N(0, 1). ---------------
#3@
# rnorm(__, __, __)
#@3
# Suppose we have this task:
# Generate 10 random numbers from N(0, 1),
# 10 random numbers from N(0, 2),
# 10 random numbers from N(0, 3),
# all the way up to 10 random numbers from N(0, 100).
# First attempt: try to put a vector into 'sd':
rnorm(n = 10, mean = 0, sd = 1:100)
# What happens? R generates one random number from N(0, 1), one random number
# from N(0, 2), one random number from N(0, 3), all the way up to one random
# number from N(0, 10). It stops at 10 because `n = 10`. `rnorm` is
# vectorized, but not in the way that helps us solve this problem.
# Second attempt: you could copy-paste 100 times, changing the sd each time:
rnorm(n = 10, mean = 0, sd = 1)
rnorm(n = 10, mean = 0, sd = 2)
rnorm(n = 10, mean = 0, sd = 3)
rnorm(n = 10, mean = 0, sd = 4)
#etc.
# The problems with the copy-paste solution:
#
# * contains a lot of extra unnecessary code for a reader to have to read
# * doesn't use the computer as the powerful tool it is
# * is annoying for you to have to write (and depending on the problem, may
# take you all day or even all week to type out!)
# The correct solution: use map(.x, .f).
# `map(.x, .f)` is from the package `purrr`, which is the last tidyverse
# package we'll talk about in-depth. `purrr` is named the way it is because
# it helps "make your functions purrr" like a well oiled machine. And that's
# exactly what we need help with in the example above: `rnorm()` wasn't
# purrring for us!
# map(.x, .f) does one simple thing: It applies the function .f to each
# element of the vector .x.
# map(.x, .f) will return a list of the same length as .x. Check out the
# diagram that explains visually how to use map():
#' 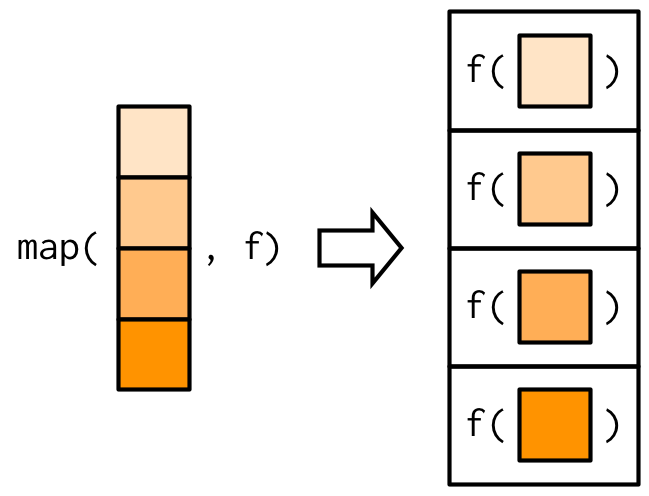
# The name `map` refers to how "map" is used in math: a mapping is an
# operation that associates each element of a set with elements in a different
# set. The `.x` are the inputs, and the `.f` is the function to apply to get
# to the set of outputs.
# So how do we use `map(.x, .f)` to solve our problem?
# I like to write the beginning of the copy-paste version:
rnorm(n = 10, mean = 0, sd = 1)
rnorm(n = 10, mean = 0, sd = 2)
rnorm(n = 10, mean = 0, sd = 3)
# What should be the vector of inputs `.x`? It's whatever needed to change in
# the code above. In that code, everything stayed the same except for the `sd`
# needed to change: it needed to go from 1 to 100.
# `.x` will be the vector 1:100.
# What's the function `.f` we'll apply to every element of `.x`? It should
# take a standard deviation as its argument, and it should output 10 ranom
# normal numbers with a mean of zero and a standard deviation that is the
# function argument:
# function(.x) {
# rnorm(n = 10, mean = 0, sd = .x)
# }
# Use the tilde `~` for defining a function "formula-style" and referring to
# .x directly in the map call:
# map(.x = 1:100, .f = ~ rnorm(n = 10, mean = 0, sd = .x))
# There are actually 3 ways to define the function `.f` inside a map call:
# * as a formula with the tilde `~`
# * as an anonymous (lambda) function
# * as a named function
# All these options are pretty similar, so it comes down to personal
# preference.
# Using an anonymous function:
# map(.x = 1:100, .f = function(input) rnorm(n = 10, mean = 0, sd = input))
# Or use a named function:
# rnorm_sd <- function(input) {
# rnorm(n = 10, mean = 0, sd = input)
# }
# map(.x = 1:100, .f = rnorm_sd)
# ----- Example 2: reading csv files -----
# Recall: in the murder mystery project, we had to read 8 different csv files,
# and we did it one by one, because 'read_csv' takes just ONE csv file at a
# time.
# people <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/person.csv")
# drivers_license <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/drivers_license.csv")
# income <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/income.csv")
# crime_scene_report <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/crime_scene_report.csv")
# facebook_event_checkin <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/facebook_event_checkin.csv")
# get_fit_now_checkin <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/get_fit_now_checkin.csv")
# get_fit_now_member <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/get_fit_now_member.csv")
# interview <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/interview.csv")
# What if you had to read 100 or 1,000 csv's? You would *not* want to type
# all that out!
# `paste()` is vectorized in just the way we want. This generates the
# character strings we need:
paste(
"https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/",
c("person", "drivers_license", "income", "crime_scene_report"),
".csv",
sep = ""
)
# But `read_csv()` is not vectorized. It takes only one csv file at a time,
# and when we pass a vector in, it thinks the names of the files are the data
# we're interested in.
paste(
"https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/",
c("person", "drivers_license", "income", "crime_scene_report"),
".csv",
sep = ""
) %>%
read_csv()
# Problem: `read_csv` doesn't purrr! Let's use `map(.x, .f)`
# .x: the things we want to change. Here, that will be the string for
# the csv file. .f: the function we want to apply to every element of `.x`.
# Here that's just 'read_csv'.
# map(
# .x = paste(
# "https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/",
# c("person", "drivers_license", "income", "crime_scene_report"),
# ".csv", sep = ""
# ),
# .f = read_csv
# )
# It works! The output is a list of tibbles. From there, we could tell R how
# to use `left_join` to combine them into a bigger tibble by their common
# variables.
#:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# Great work! You're one step closer to tidyverse enlightenment.
# Make sure to return to this topic to meditate on it later.
# If you're ready, you can move on to the next koan: more map().
}
## function() {#:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
## # Intro to the Tidyverse by Colleen O'Briant
## # Koan #15: map()
## #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
##
## # In order to progress:
## # 1. Read all instructions carefully.
## # 2. When you come to an exercise, fill in the blank, un-comment the line
## # (Ctrl/Cmd Shift C), and execute the code in the console (Ctrl/Cmd Return).
## # If the piece of code spans multiple lines, highlight the whole chunk or
## # simply put your cursor at the end of the last line.
## # 3. Save (Ctrl/Cmd S).
## # 4. Test that your answers are correct (Ctrl/Cmd Shift T).
##
## #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
##
## # In this class, we've learned a lot about two very important tidyverse
## # packages: 'dplyr' and 'ggplot2'. We'll spend the rest of the course
## # learning a third: 'purrr'. We'll only learn 3 functions from that
## # package, but they are deep concepts and really powerful tools. The
## # three functions are map(), reduce(), and accumulate().
##
## # This koan introduces map(), the next one gives you more practice using
## # it, and reduce() and accumulate() will come a little later in koans
## # 18-20.
##
## # Run this code to get started:
##
## library(tidyverse)
## library(gapminder)
##
## #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
##
## # ----- Vectorized Functions -----
##
## # Last week we learned we could define our own custom functions. Here I
## # define a function called 'pct_change' that takes two arguments: an "old"
## # value and a "new" value, and it returns the percentage change between
## # them.
##
## pct_change <- function(old, new) {
## (new - old) / old
## }
##
## # 1. Does 'pct_change' work on values? ---------------------------------------
##
## #1@
##
## # pct_change(__, __) == 1
##
## #@1
##
##
## # 2. Does 'pct_change' work on vectors? --------------------------------------
##
## #2@
##
## # pct_change(__, __) == c(0, 1, 2)
##
## #@2
##
##
## # How is it that 'pct_change' works on vectors? Well, 'pct_change' is defined
## # only using subtraction and division, which both work element-wise on
## # vectors. You can plug vectors directly into the computation that's done in
## # the function body:
##
## (c(4, 5, 6) - c(1, 2, 3)) / c(1, 2, 3)
##
## # ----- Example 1: rnorm -----
##
## # Most of the functions we've been using are "vectorized" (they work with
## # vectors of any length). But some functions may not work on vectors so
## # smoothly. And when you define your own custom functions that are more
## # complicated than `pct_change`, they oftentimes won’t work on vectors.
##
## # For example, take 'rnorm()'.
##
## # 3. Use 'rnorm()' to generate 10 random numbers from N(0, 1). ---------------
##
## #3@
##
## # rnorm(__, __, __)
##
## #@3
##
##
## # Suppose we have this task:
## # Generate 10 random numbers from N(0, 1),
## # 10 random numbers from N(0, 2),
## # 10 random numbers from N(0, 3),
## # all the way up to 10 random numbers from N(0, 100).
##
## # First attempt: try to put a vector into 'sd':
## rnorm(n = 10, mean = 0, sd = 1:100)
##
## # What happens? R generates one random number from N(0, 1), one random number
## # from N(0, 2), one random number from N(0, 3), all the way up to one random
## # number from N(0, 10). It stops at 10 because `n = 10`. `rnorm` is
## # vectorized, but not in the way that helps us solve this problem.
##
## # Second attempt: you could copy-paste 100 times, changing the sd each time:
## rnorm(n = 10, mean = 0, sd = 1)
## rnorm(n = 10, mean = 0, sd = 2)
## rnorm(n = 10, mean = 0, sd = 3)
## rnorm(n = 10, mean = 0, sd = 4)
## #etc.
##
## # The problems with the copy-paste solution:
## #
## # * contains a lot of extra unnecessary code for a reader to have to read
## # * doesn't use the computer as the powerful tool it is
## # * is annoying for you to have to write (and depending on the problem, may
## # take you all day or even all week to type out!)
##
##
## # The correct solution: use map(.x, .f).
##
## # `map(.x, .f)` is from the package `purrr`, which is the last tidyverse
## # package we'll talk about in-depth. `purrr` is named the way it is because
## # it helps "make your functions purrr" like a well oiled machine. And that's
## # exactly what we need help with in the example above: `rnorm()` wasn't
## # purrring for us!
##
## # map(.x, .f) does one simple thing: It applies the function .f to each
## # element of the vector .x.
##
## # map(.x, .f) will return a list of the same length as .x. Check out the
## # diagram that explains visually how to use map():
##
## #' 
##
## # The name `map` refers to how "map" is used in math: a mapping is an
## # operation that associates each element of a set with elements in a different
## # set. The `.x` are the inputs, and the `.f` is the function to apply to get
## # to the set of outputs.
##
## # So how do we use `map(.x, .f)` to solve our problem?
##
##
## # I like to write the beginning of the copy-paste version:
## rnorm(n = 10, mean = 0, sd = 1)
## rnorm(n = 10, mean = 0, sd = 2)
## rnorm(n = 10, mean = 0, sd = 3)
##
## # What should be the vector of inputs `.x`? It's whatever needed to change in
## # the code above. In that code, everything stayed the same except for the `sd`
## # needed to change: it needed to go from 1 to 100.
##
## # `.x` will be the vector 1:100.
##
## # What's the function `.f` we'll apply to every element of `.x`? It should
## # take a standard deviation as its argument, and it should output 10 ranom
## # normal numbers with a mean of zero and a standard deviation that is the
## # function argument:
##
## # function(.x) {
## # rnorm(n = 10, mean = 0, sd = .x)
## # }
##
## # Use the tilde `~` for defining a function "formula-style" and referring to
## # .x directly in the map call:
##
## # map(.x = 1:100, .f = ~ rnorm(n = 10, mean = 0, sd = .x))
##
## # There are actually 3 ways to define the function `.f` inside a map call:
##
## # * as a formula with the tilde `~`
## # * as an anonymous (lambda) function
## # * as a named function
##
## # All these options are pretty similar, so it comes down to personal
## # preference.
##
## # Using an anonymous function:
##
## # map(.x = 1:100, .f = function(input) rnorm(n = 10, mean = 0, sd = input))
##
## # Or use a named function:
##
## # rnorm_sd <- function(input) {
## # rnorm(n = 10, mean = 0, sd = input)
## # }
## # map(.x = 1:100, .f = rnorm_sd)
##
##
## # ----- Example 2: reading csv files -----
##
## # Recall: in the murder mystery project, we had to read 8 different csv files,
## # and we did it one by one, because 'read_csv' takes just ONE csv file at a
## # time.
##
## # people <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/person.csv")
## # drivers_license <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/drivers_license.csv")
## # income <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/income.csv")
## # crime_scene_report <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/crime_scene_report.csv")
## # facebook_event_checkin <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/facebook_event_checkin.csv")
## # get_fit_now_checkin <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/get_fit_now_checkin.csv")
## # get_fit_now_member <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/get_fit_now_member.csv")
## # interview <- read_csv("https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/interview.csv")
##
## # What if you had to read 100 or 1,000 csv's? You would *not* want to type
## # all that out!
##
## # `paste()` is vectorized in just the way we want. This generates the
## # character strings we need:
##
## paste(
## "https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/",
## c("person", "drivers_license", "income", "crime_scene_report"),
## ".csv",
## sep = ""
## )
##
## # But `read_csv()` is not vectorized. It takes only one csv file at a time,
## # and when we pass a vector in, it thinks the names of the files are the data
## # we're interested in.
##
## paste(
## "https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/",
## c("person", "drivers_license", "income", "crime_scene_report"),
## ".csv",
## sep = ""
## ) %>%
## read_csv()
##
## # Problem: `read_csv` doesn't purrr! Let's use `map(.x, .f)`
##
## # .x: the things we want to change. Here, that will be the string for
## # the csv file. .f: the function we want to apply to every element of `.x`.
## # Here that's just 'read_csv'.
##
## # map(
## # .x = paste(
## # "https://raw.githubusercontent.com/cobriant/dplyrmurdermystery/master/data/",
## # c("person", "drivers_license", "income", "crime_scene_report"),
## # ".csv", sep = ""
## # ),
## # .f = read_csv
## # )
##
## # It works! The output is a list of tibbles. From there, we could tell R how
## # to use `left_join` to combine them into a bigger tibble by their common
## # variables.
##
## #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
##
## # Great work! You're one step closer to tidyverse enlightenment.
## # Make sure to return to this topic to meditate on it later.
##
## # If you're ready, you can move on to the next koan: more map().
## }